The gamma distribution is a continuous probability distribution that is popular for a range of phylogenetic applications. The gamma distribution is popular in part because its a bit of a shape shifter that can assume a range of shapes, from exponential to normal. This flexibility results from the fact that gamma distribution has two parameters. In most phylogenetic applications, these parameters are referred to as the shape parameter, , and the rate parameter, , (note that some applications of the rate parameter, , parameter is replaced by the “scale parameter,” which is simply the inverse of the rate parameter).
You can explore the impact of these two parameters on the gamma distribution using the following R scripts (note that this script focuses on plotting values that flank the default parameter settings for the gamma distribution in the program SIMMAP 1.5):
Using the R script below, you can generate probability densities for the gamma distribution that vary the value of the shape parameter () (Fig. 1). As you can see, varying has a strong impact on the shape of the gamma distribution. The gamma distribution is the sum of independent and identically distributed (i.i.d.) exponential distributions (i.e., that have the same rate parameter). Accordingly, when = 1, the gamma collapses to an exponential distribution, when >> 1, the gamma distribution increasingly resembles a normal distribution.
Figure 1: The impact of varying the shape parameter (alpha) on the gamma distribution.
#Generate a plot of gamma distributions that vary the shape parameter (alpha).
x <- seq(0, 100, length=200)
simmapDefaultGamma <- dgamma(x, shape=1.25, scale=1/0.25) #Make probability density function for SIMMAP default gamma distribution
plot(x, simmapDefaultGamma, type="l", yaxs="i", xaxs="i", ylim=c(0,0.16), xlim=c(0,100), xlab="x value", ylab="Density", main="Probability density for gamma distribution with variable alpha and beta=0.25", lwd=0)
colors <- c("red", "black", "blue", "darkgreen", "purple", "orange")
alphas <- c(0.1, 1.25, 2, 4, 8, 10)
labels <- c("alpha=0.1", "alpha=1.25 (SIMMAP default)", "alpha=2", "alpha=4", "alpha=8", "alpha=10")
for(i in 1:length(alphas)) {
hx <- dgamma(x, shape=alphas[i], rate=1, scale=1/0.25)
lines(x, hx, lwd=3, col=colors[i])
}
legend("topright", inset=.05, title="Probability densities",
labels, lwd=3, col=colors)
Using the R script below, you can visualize the impact of varying the rate parameter () while keeping the shape parameter () constant (Fig. 2). has a strong impact on the shape of the gamma distribution. When is set to less than 1, we tend to observe relatively broad distributions with long tails. As we increase the value of , we observe increasingly tight distributions. This effect stems from the fact that the variance of the gamma is /.
Figure 2: The impact of varying the rate parameter (beta) on the gamma distribution.
#Generate a plot of gamma distributions that vary the rate parameter (beta) as in Figure 2 below.
x <- seq(0, 100, length=200)
simmapDefaultGamma <- dgamma(x, shape=1.25, scale=1/0.25) #Make probability density function for SIMMAP default gamma distribution
#plot(x, simmapDefaultGamma, type="l")
plot(x, simmapDefaultGamma, type="l", yaxs="i", xaxs="i", ylim=c(0,0.9), xlim=c(0,70), xlab="x value", ylab="Density", main="Probability density for gamma distribution with a fixed alpha=1.25 and variable beta", lwd=2)
colors <- c("red", "black", "blue", "darkgreen", "purple", "orange")
betas <- c(0.1, 0.25, 2, 4, 8, 10)
labels <- c("beta=0.1", "beta=0.25 (SIMMAP default)", "beta=2", "beta=4", "beta=8", "beta=10")
for(i in 1:length(betas)) {
hx <- dgamma(x, shape=1.25, rate=1, scale=1/betas[i])
lines(x, hx, lwd=2, col=colors[i])
}
legend("topright", inset=.05, title="Probability densities",labels, lwd=2, col=colors)
For many phylogenetic applications of the gamma distribution -- e.g, to accommodate variation in substitution rate across sites (ASRV) -- the and parameters are constrained to be equal. You can visualize the gamma distributions generated under these conditions using the R script below (Fig. 3). Because the mean of the gamma distribution is , this constraint ensures that the gamma distribution has a mean of one. This is important when the gamma distribution is used as a prior probability density on ASRV, as it retains the ability to interpret branch lengths as the expected (mean) number of substitutions per site.
Figure 3: Gamma distributions with alpha and beta set equal to one another.
#Generate a plot of gamma distributions with alpha and beta equal to one another as in Figure 3 below.
x <- seq(0, 100, length=200)
simmapDefaultGamma <- dgamma(x, shape=1.25, scale=1/0.25) #Make probability density function for SIMMAP default gamma distribution
#plot(x, simmapDefaultGamma, type="l")
plot(x, simmapDefaultGamma, type="l", yaxs="i", xaxs="i", ylim=c(0,2), xlim=c(0,30), xlab="x value", ylab="Density", main="Probability density for gamma distribution with a alpha and beta equal to one another", lwd=2)
colors <- c("red", "black", "blue", "darkgreen", "purple", "orange")
alphas <- c(0.1, 0.5, 1, 5, 20)
betas <- c(0.1, 0.5, 1, 5, 20)
labels <- c("alpha=0.1, beta=0.1", "alpha=1.25, beta=0.25 (SIMMAP default)", "alpha=0.5, beta=0.5", "alpha=1, beta=1", "alpha=5, beta=5", "alpha=20, beta=20")
for(i in 1:length(betas)) {
hx <- dgamma(x, shape=alphas[i], rate=1, scale=1/betas[i])
lines(x, hx, lwd=2, col=colors[i])
}
legend("topright", inset=.05, title="Probability densities",labels, lwd=2, col=colors)
Great idea for a post. Personally, I find that playing around with probability functions in R is the best way to develop an intuition for them.
This bit of code should drive home the point that the sum of k exponential random variable with rate &lambda is gamma distributed with &alpha = k and &beta = &lambda.
k = 1000
lambda = 1
# simulate the sum of k exponential(1) random variables 10000 times.
x = sapply(1:10000,function(x) sum(rexp(n=k,rate=lambda)))
# plot the histogram of the simulated sums.
hist(x,freq=FALSE,breaks=50)
# plot the probability density of the gamma distribution with alpha = k and beta = lambda.
curve(dgamma(x=x,shape=k,rate=lambda),from=min(x),to=max(x),n=1000,add=TRUE)
(Apologies in advance if my tags are messed up, there is no preview!)
I agree that this could be the start of a nice `probability distributions in R’ series (so feel free to dive in!). This would be useful in general, and a nice primer for the Bodega workshop.
As an (obscure) aside about the mean-one gamma: one of its many uses is as a prior probability density for rate multipliers in the compound Poisson process (CPP) relaxed-clock model proposed by Huelsenbeck et al. (2000)—substitution-rate shifts occur along the tree as Poisson events (with exponentially distributed waiting times, with rate ), and when they occur, the current rate is multiplied by a gamma-distributed random variable. When I originally read the CPP paper, I thought that (in keeping with the logic of the use of the mean-one gamma for accommodating ASRV), that the rate multipliers were also mean-one gamma distributed random variable .
However, it turns out that the product of mean-one gamma-distributed multipliers converges to zero as gets large. Accordingly, Huelsenbeck et al. (2000) actually parameterize the gamma prior on rate multipliers so that the mean of the log of the rate multipliers is zero: The rate multipliers are therefore approximately mean-one gamma-distributed random variables: , where is the derivative of the log of the gamma function.—Brian
I think it is worth reminding people that in SIMMAP the gamma distribution is only used for the overall substitution rate of a character while a symmetric beta prior is used for the morphological state frequencies.
In trying to understand the gamma distribution, interactive plots are really useful.
Shiny lets you make fully interactive data reports in the browser.
ggvis lets you make plots interactive using javascript, but is its own plotting paradigm (similar to ggplot2).
One more is manipulate, which requires rstudio as far as I know.
But in rstudio, you can run:
library(manipulate)
library(ggplot2)
manipulate({qplot(x=x,y=dgamma(x,shape=s,rate=r),geom='line') + scale_y_continuous(limit=c(0,1)) + scale_x_continuous(limit=c(0,100)) + theme_classic()},s=slider(0,100,step=.1),r=slider(0,100,step=.1))
At some point, I am going to work on shiny apps for visualizing distributions. I had a few, and even had a power analysis tool, but I just recreated my own webpage and those are not yet running.
Great idea for a post. Personally, I find that playing around with probability functions in R is the best way to develop an intuition for them.
This bit of code should drive home the point that the sum of k exponential random variable with rate &lambda is gamma distributed with &alpha = k and &beta = &lambda.
k = 1000
lambda = 1
# simulate the sum of k exponential(1) random variables 10000 times.
x = sapply(1:10000,function(x) sum(rexp(n=k,rate=lambda)))
# plot the histogram of the simulated sums.
hist(x,freq=FALSE,breaks=50)
# plot the probability density of the gamma distribution with alpha = k and beta = lambda.
curve(dgamma(x=x,shape=k,rate=lambda),from=min(x),to=max(x),n=1000,add=TRUE)
(Apologies in advance if my tags are messed up, there is no preview!)
Timely post! I was just explaining this the other day and wishing I had some R code handy to demonstrate. Thanks guys.
Mike, sorry about the tags. We have convenient latex support, although this is the first time I’ve tried it in a comment. Let’s see how it looks.
To use it, use latex tags as follows
$latexyour-code-here$I agree that this could be the start of a nice `probability distributions in R’ series (so feel free to dive in!). This would be useful in general, and a nice primer for the Bodega workshop.
As an (obscure) aside about the mean-one gamma: one of its many uses is as a prior probability density for rate multipliers in the compound Poisson process (CPP) relaxed-clock model proposed by Huelsenbeck et al. (2000)—substitution-rate shifts occur along the tree as Poisson events (with exponentially distributed waiting times, with rate ), and when they occur, the current rate is multiplied by a gamma-distributed random variable. When I originally read the CPP paper, I thought that (in keeping with the logic of the use of the mean-one gamma for accommodating ASRV), that the rate multipliers were also mean-one gamma distributed random variable
), and when they occur, the current rate is multiplied by a gamma-distributed random variable. When I originally read the CPP paper, I thought that (in keeping with the logic of the use of the mean-one gamma for accommodating ASRV), that the rate multipliers were also mean-one gamma distributed random variable  .
.
However, it turns out that the product of mean-one gamma-distributed multipliers converges to zero as gets large. Accordingly, Huelsenbeck et al. (2000) actually parameterize the gamma prior on rate multipliers so that the mean of the log of the
gets large. Accordingly, Huelsenbeck et al. (2000) actually parameterize the gamma prior on rate multipliers so that the mean of the log of the 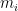 rate multipliers is zero:
rate multipliers is zero: ![\mathbb{E}[\log(m_i)] = 0. \mathbb{E}[\log(m_i)] = 0.](//s0.wp.com/latex.php?latex=%5Cmathbb%7BE%7D%5B%5Clog%28m_i%29%5D+%3D+0.&bg=ffffff&fg=000&s=0) The rate multipliers are therefore approximately mean-one gamma-distributed random variables:
The rate multipliers are therefore approximately mean-one gamma-distributed random variables: 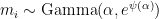 , where
, where 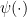 is the derivative of the log of the gamma function.—Brian
is the derivative of the log of the gamma function.—Brian
I think it is worth reminding people that in SIMMAP the gamma distribution is only used for the overall substitution rate of a character while a symmetric beta prior is used for the morphological state frequencies.
Pingback: Jukes Cantor Model of DNA substitution | Workshop in Applied Phylogenetics
In trying to understand the gamma distribution, interactive plots are really useful.
Shiny lets you make fully interactive data reports in the browser.
ggvis lets you make plots interactive using javascript, but is its own plotting paradigm (similar to ggplot2).
One more is manipulate, which requires rstudio as far as I know.
But in rstudio, you can run:
library(manipulate)
library(ggplot2)
manipulate({qplot(x=x,y=dgamma(x,shape=s,rate=r),geom='line') + scale_y_continuous(limit=c(0,1)) + scale_x_continuous(limit=c(0,100)) + theme_classic()},s=slider(0,100,step=.1),r=slider(0,100,step=.1))
At some point, I am going to work on shiny apps for visualizing distributions. I had a few, and even had a power analysis tool, but I just recreated my own webpage and those are not yet running.